R编程中的置换假设检验
简单来说,R中的置换假设检验是一种比较两组数值的方法。置换假设检验可以替代:
- 独立二样本 t 检验
- Mann-Whitney U 又名 Wilcoxon 秩和检验
让我们在 R 编程中实现这个测试。
为什么要使用置换假设检验?
- 小样本量。
- 不满足假设(对于参数方法)。
- 测试比较均值和中位数的经典方法以外的其他方法。
- 难以估计检验统计量的 SE。
置换假设检验步骤
- 指定假设
- 选择 test-stat(例如:Mean、Median 等)
- 确定 test-stat 的分布
- 将 test-stat 转换为 P 值
Note:
P-value = No. of permutations having a test-stat value greater than observed test-stat value/ No. of permutations.
在 R 中的实现
- 数据集:鸡肉饮食数据。该数据集是“R 数据集包”中“chickwts”数据的子集。在此处下载数据集。
- 假设:鸡的重量与饮食类型无关。
检验统计
- 检验统计 #1:两种饮食的平均体重差异的绝对值| Y 1 – Y 2 | .这是与独立的两侧双样本t 检验相同的检验统计量。
- 检验统计 #2:两种饮食的中位数体重差异的绝对值|中位数1 – 中位数2 |
R
# R program to illustrate
# Permutation Hypothesis Test
# load the data set
d <- read.table(file = "ChickData.csv",
header = T, sep = ",")
# print the dataset
print(d)
# check the names
names(d)
levels(d$feed)
# how many observations in each diet?
table(d$feed)
# let's look at a boxplot of weight gain by those 2 diets
boxplot(d$weight~d$feed, las = 1,
ylab = "weight (g)",
xlab = "feed",
main = "Weight by Feed")
# calculate the difference in sample MEANS
mean(d$weight[d$feed == "casein"]) # mean for casein
mean(d$weight[d$feed == "meatmeal"]) # mean for meatmeal
# lets calculate the absolute diff in means
test.stat1 <- abs(mean(d$weight[d$feed == "casein"]) -
mean(d$weight[d$feed == "meatmeal"]))
test.stat1
# calculate the difference in sample MEDIANS
median(d$weight[d$feed == "casein"]) # median for casein
median(d$weight[d$feed == "meatmeal"]) # median for meatmeal
# lets calculate the absolute diff in medians
test.stat2 <- abs(median(d$weight[d$feed == "casein"]) -
median(d$weight[d$feed == "meatmeal"]))
test.stat2
# Permutation Test
# for reproducability of results
set.seed(1979)
# the number of observations to sample
n <- length(d$feed)
# the number of permutation samples to take
P <- 100000
# the variable we will resample from
variable <- d$weight
# initialize a matrix to store the permutation data
PermSamples <- matrix(0, nrow = n, ncol = P)
# each column is a permutation sample of data
# now, get those permutation samples, using a loop
# let's take a moment to discuss what that code is doing
for(i in 1:P)
{
PermSamples[, i] <- sample(variable,
size = n,
replace = FALSE)
}
# we can take a quick look at the first 5 columns of PermSamples
PermSamples[, 1:5]
# initialize vectors to store all of the Test-stats
Perm.test.stat1 <- Perm.test.stat2 <- rep(0, P)
# loop thru, and calculate the test-stats
for (i in 1:P)
{
# calculate the perm-test-stat1 and save it
Perm.test.stat1[i] <- abs(mean(PermSamples[d$feed == "casein",i]) -
mean(PermSamples[d$feed == "meatmeal",i]))
# calculate the perm-test-stat2 and save it
Perm.test.stat2[i] <- abs(median(PermSamples[d$feed == "casein",i]) -
median(PermSamples[d$feed == "meatmeal",i]))
}
# before going too far with this,
# let's remind ourselves of
# the TEST STATS
test.stat1; test.stat2
# and, take a look at the first 15
# permutation-TEST STATS for 1 and 2
round(Perm.test.stat1[1:15], 1)
round(Perm.test.stat2[1:15], 1)
# and, let's calculate the permutation p-value
# notice how we can ask R a true/false question
(Perm.test.stat1 >= test.stat1)[1:15]
# and if we ask for the mean of all of those,
# it treats 0 = FALSE, 1 = TRUE
mean((Perm.test.stat1 >= test.stat1)[1:15])
# Calculate the p-value, for all P = 100,000
mean(Perm.test.stat1 >= test.stat1)
# and, let's calculate the p-value for
# option 2 of the test statistic (abs diff in medians)
mean(Perm.test.stat2 >= test.stat2)输出:
> print(d)
weight feed
1 325 meatmeal
2 257 meatmeal
3 303 meatmeal
4 315 meatmeal
5 380 meatmeal
6 153 meatmeal
7 263 meatmeal
8 242 meatmeal
9 206 meatmeal
10 344 meatmeal
11 258 meatmeal
12 368 casein
13 390 casein
14 379 casein
15 260 casein
16 404 casein
17 318 casein
18 352 casein
19 359 casein
20 216 casein
21 222 casein
22 283 casein
23 332 casein
> names(d)
[1] "weight" "feed"
> levels(d$feed)
[1] "casein" "meatmeal"
> table(d$feed)
casein meatmeal
12 11 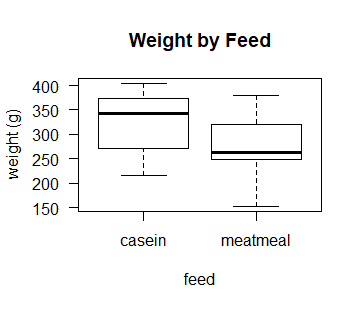
> mean(d$weight[d$feed == "casein"]) # mean for casein
[1] 323.5833
> mean(d$weight[d$feed == "meatmeal"]) # mean for meatmeal
[1] 276.9091
> test.stat1
[1] 46.67424
> median(d$weight[d$feed == "casein"]) # median for casein
[1] 342
> median(d$weight[d$feed == "meatmeal"]) # median for meatmeal
[1] 263
> test.stat2
[1] 79
> PermSamples[, 1:5]
[,1] [,2] [,3] [,4] [,5]
[1,] 379 283 380 352 206
[2,] 380 303 258 260 380
[3,] 257 206 379 380 153
[4,] 283 242 222 404 359
[5,] 222 260 325 258 258
[6,] 315 352 153 379 263
[7,] 352 263 263 325 325
[8,] 153 325 315 359 216
[9,] 368 379 344 242 260
[10,] 344 258 368 368 257
[11,] 359 257 206 257 315
[12,] 206 153 404 222 303
[13,] 404 344 303 390 390
[14,] 325 318 318 303 352
[15,] 242 404 332 263 404
[16,] 390 380 257 206 379
[17,] 260 332 216 315 318
[18,] 303 359 352 344 368
[19,] 263 222 242 283 222
[20,] 332 368 260 332 344
[21,] 318 315 283 318 283
[22,] 216 390 390 153 332
[23,] 258 216 359 216 242
> test.stat1; test.stat2
[1] 46.67424
[1] 79
> round(Perm.test.stat1[1:15], 1)
[1] 17.1 32.4 17.6 47.1 56.1 28.9 31.0 40.8 6.8 13.8 9.1 46.5 28.9 50.9 32.7
> round(Perm.test.stat2[1:15], 1)
[1] 61.0 75.0 4.5 59.0 78.0 17.0 62.0 38.5 4.5 16.0 23.0 60.5 63.5 75.0 37.0
> (Perm.test.stat1 >= test.stat1)[1:15]
[1] FALSE FALSE FALSE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE TRUE FALSE
> mean((Perm.test.stat1 >= test.stat1)[1:15])
[1] 0.2
> mean(Perm.test.stat1 >= test.stat1)
[1] 0.09959
> mean(Perm.test.stat2 >= test.stat2)
[1] 0.05407